Function to Extract Planet Labs Data by Parcel in R

The following code can be copied into R and ran as the function “PlanetToNDVI”. You can change the band math and function name to produce any index you wish, or a single band. The output is a data frame representing administrative boundaries and a column containing the average index values from Planet Labs data. Note that this requires 2 files in your root directory: 1.) 1 or many Planet labs products and a shapefile representing some sort of administrative boundary (Parcels, Census Blocks, Counties etc.).
###Prelims
library(tmap)
library(mapview)
library(sf)
library(raster)
library(stringr)
library(questionr)
library(dplyr)
library(rgdal)
library(rgeos)
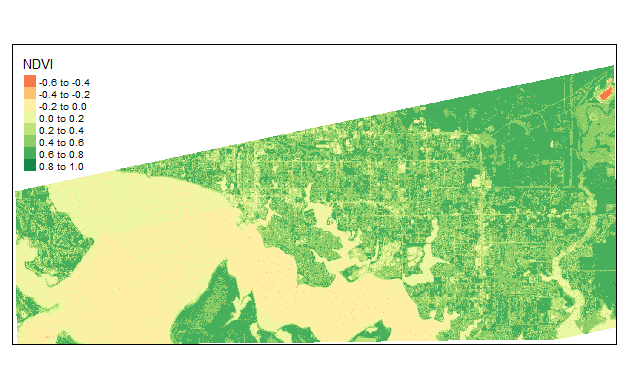
###Single image example
PlanetImage<- stack("Data/10_12_2018_PSScene4Band_Explorer/files/20181012_154650_1_104a_3B_AnalyticMS_SR_clip.tif") #Upload 4band Image
ndvi <- (PlanetImage[[4]] - PlanetImage[[1]])/(PlanetImage[[4]] + PlanetImage[[1]]) #make NDVI
tmap_mode("view")
tm_shape(ndvi) +
tm_raster( title = "NDVI")
###Extract Average Index Value by Parcel as Function
PlanetToNDVI<-function(ImageLocation,FileOutput,ImageDate,AssetType,UOA){
# make list of file names that are rasters. Note to change the pattern argument if rasters are not .tif. Alternatively you can remove the pattern argument if the file location only stores relevant raster files to be mosaiced.
current.list <- list.files(path=paste(ImageLocation,ImageDate,"_",AssetType,"_Explorer/files/", sep =""),
pattern ="SR_clip.tif$",
full.names=TRUE)
# read in raster files as a raster stack
images<- lapply(current.list, stack)
# clear user defined names if present
names(images) <- NULL
# this tells the mosaic function to average any overlapping pixels
raster.list$fun <- mean
# mosaic list using the mosaic function the raster package
imageMosaic <- do.call(mosaic, raster.list)
#calculate index
ndvi <- (imageMosaic[[4]] - imageMosaic[[3]])/(imageMosaic[[4]] + imageMosaic[[3]])
Parcels = read_sf(UOA) #Load in its of analysis shapefile and reproject image coordinate system
Parcels<-st_transform(Parcels, crs(imageMosaic))
ndvi[ndvi < 0.2] <- NA #mask off index below threshold to try and omit roof, driveway etc. Consider deriving threshold from ML algo at some point...
ex <- extract(ndvi,
Parcels,
fun=mean,
na.rm=TRUE,
df=TRUE)
ex$GlobalID <-Parcels$GlobalID
names(ex)[2] <- ImageDate
write.csv(ex, paste(FileOutput,ImageDate,".csv", sep=""))
}
Run Function
PlanetToNDVI("Data/",
"Ouputs/",
"10_12_2018",
"PSScene4Band",
"ParcelAOI.shp")